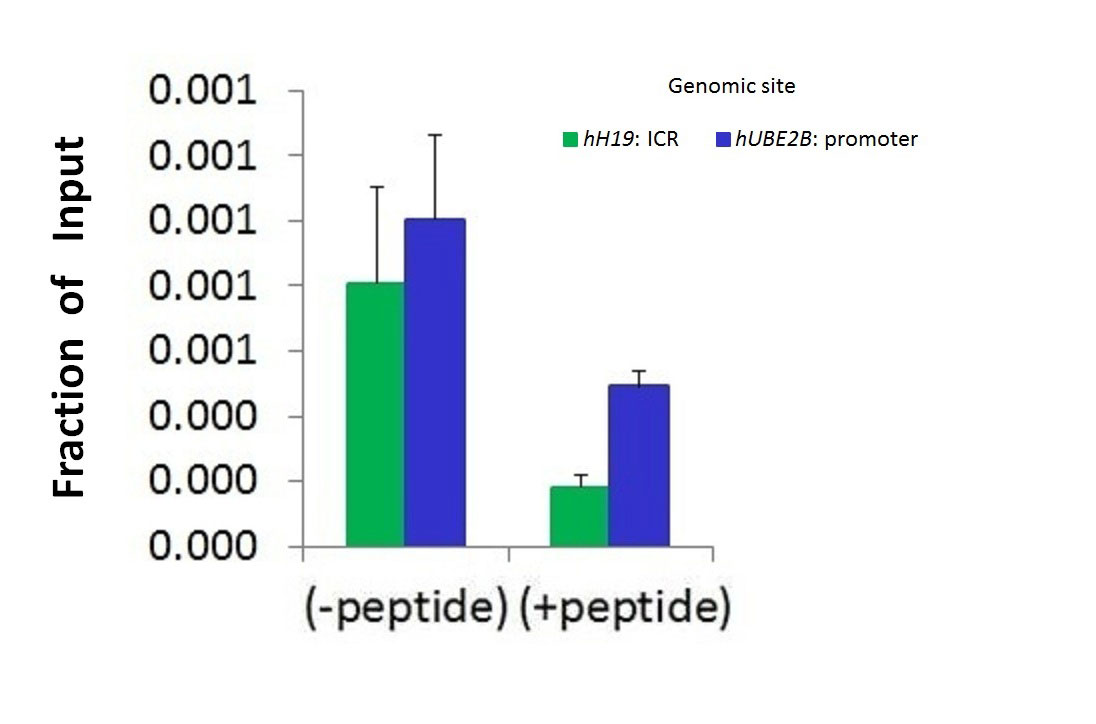
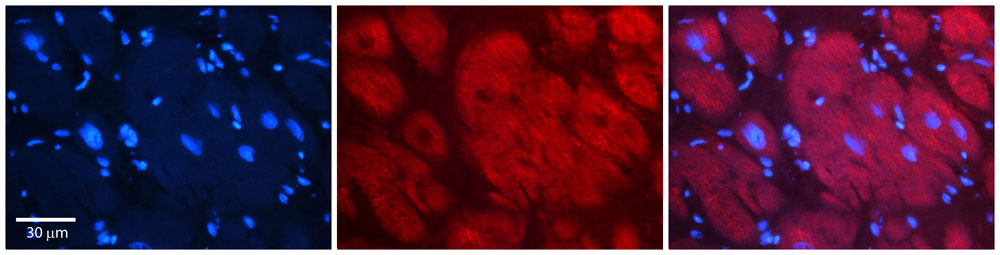
Hdac6 Antibody - C-terminal region
Rabbit Polyclonal Antibody
- 产品详情
- 实验流程
Application
| WB, CHIP |
|---|---|
| Primary Accession | Q9Z2V5 |
| Other Accession | NM_010413, NP_034543 |
| Reactivity | Human, Mouse, Rat, Rabbit, Pig, Dog, Horse, Bovine |
| Predicted | Human, Mouse, Rat, Pig, Dog, Bovine |
| Host | Rabbit |
| Clonality | Polyclonal |
| Calculated MW | 125787 Da |
| Gene ID | 15185 |
|---|---|
| Alias Symbol | Hd6, Hdac5, Sfc6, mHDA2 |
| Other Names | Histone deacetylase 6, HD6, 3.5.1.98, Histone deacetylase mHDA2, Hdac6 |
| Format | Liquid. Purified antibody supplied in 1x PBS buffer with 0.09% (w/v) sodium azide and 2% sucrose. |
| Reconstitution & Storage | Add 50 ul of distilled water. Final anti-Hdac6 antibody concentration is 1 mg/ml in PBS buffer with 2% sucrose. For longer periods of storage, store at 20°C. Avoid repeat freeze-thaw cycles. |
| Precautions | Hdac6 Antibody - C-terminal region is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | Hdac6 {ECO:0000312|MGI:MGI:1333752} |
|---|---|
| Function | Deacetylates a wide range of non-histone substrates (PubMed:12606581, PubMed:19893491, PubMed:26746851, PubMed:27737934). Plays a central role in microtubule-dependent cell motility by mediating deacetylation of tubulin (PubMed:19893491, PubMed:27737934, PubMed:12606581). Required for cilia disassembly via deacetylation of alpha-tubulin (By similarity). Alpha-tubulin deacetylation results in destabilization of dynamic microtubules (PubMed:12486003). Promotes deacetylation of CTTN, leading to actin polymerization, promotion of autophagosome-lysosome fusion and completion of autophagy (By similarity). Deacetylates SQSTM1 (By similarity). Deacetylates peroxiredoxins PRDX1 and PRDX2, decreasing their reducing activity (By similarity). Deacetylates antiviral protein RIGI in the presence of viral mRNAs which is required for viral RNA detection by RIGI (PubMed:26746851). Sequentially deacetylates and polyubiquitinates DNA mismatch repair protein MSH2 which leads to MSH2 degradation, reducing cellular sensitivity to DNA-damaging agents and decreasing cellular DNA mismatch repair activities (By similarity). Deacetylates DNA mismatch repair protein MLH1 which prevents recruitment of the MutL alpha complex (formed by the MLH1-PMS2 heterodimer) to the MutS alpha complex (formed by the MSH2-MSH6 heterodimer), leading to tolerance of DNA damage (By similarity). Deacetylates RHOT1/MIRO1 which blocks mitochondrial transport and mediates axon growth inhibition (By similarity). Deacetylates transcription factor SP1 which leads to increased expression of ENG, positively regulating angiogenesis (By similarity). Deacetylates KHDRBS1/SAM68 which regulates alternative splicing by inhibiting the inclusion of CD44 alternate exons (By similarity). Promotes odontoblast differentiation following IPO7- mediated nuclear import and subsequent repression of RUNX2 expression (PubMed:35922041). In addition to its protein deacetylase activity, plays a key role in the degradation of misfolded proteins: when misfolded proteins are too abundant to be degraded by the chaperone refolding system and the ubiquitin-proteasome, mediates the transport of misfolded proteins to a cytoplasmic juxtanuclear structure called aggresome (By similarity). Probably acts as an adapter that recognizes polyubiquitinated misfolded proteins and target them to the aggresome, facilitating their clearance by autophagy (PubMed:22819792). |
| Cellular Location | Cytoplasm. Cytoplasm, cytoskeleton. Nucleus. Perikaryon. Cell projection, dendrite. Cell projection, axon. Cell projection, cilium {ECO:0000250|UniProtKB:Q9UBN7}. Cytoplasm, cytoskeleton, microtubule organizing center, centrosome {ECO:0000250|UniProtKB:Q9UBN7} Cytoplasm, cytoskeleton, cilium basal body {ECO:0000250|UniProtKB:Q9UBN7}. Note=Mainly cytoplasmic where it is associated with microtubules (PubMed:10873806, PubMed:19893491). Can shuttle between the cytoplasm and the nucleus (PubMed:10873806). Found exclusively in the cytoplasm in proliferative cells with a fraction found in the nucleus during differentiation (PubMed:10873806) |
| Tissue Location | Expressed in neurons of the cortex. Expressed in Purkinje cells. Detected in keratinocytes (at protein level) |
Research Areas
For Research Use Only. Not For Use In Diagnostic Procedures.
Application Protocols
Provided below are standard protocols that you may find useful for product applications.
终于等到您。ABCEPTA(百远生物)抗体产品。
点击下方“我要评价 ”按钮提交您的反馈信息,您的反馈和评价是我们最宝贵的财富之一,
我们将在1-3个工作日内处理您的反馈信息。
如有疑问,联系:0512-88856768 tech-china@abcepta.com.























 癌症的基本特征包括细胞增殖、血管生成、迁移、凋亡逃避机制和细胞永生等。找到癌症发生过程中这些通路的关键标记物和对应的抗体用于检测至关重要。
癌症的基本特征包括细胞增殖、血管生成、迁移、凋亡逃避机制和细胞永生等。找到癌症发生过程中这些通路的关键标记物和对应的抗体用于检测至关重要。 为您推荐一个泛素化位点预测神器——泛素化分析工具,可以为您的蛋白的泛素化位点作出预测和评分。
为您推荐一个泛素化位点预测神器——泛素化分析工具,可以为您的蛋白的泛素化位点作出预测和评分。 细胞自噬受体图形绘图工具为你的蛋白的细胞受体结合位点作出预测和评分,识别结合到自噬通路中的蛋白是非常重要的,便于让我们理解自噬在正常生理、病理过程中的作用,如发育、细胞分化、神经退化性疾病、压力条件下、感染和癌症。
细胞自噬受体图形绘图工具为你的蛋白的细胞受体结合位点作出预测和评分,识别结合到自噬通路中的蛋白是非常重要的,便于让我们理解自噬在正常生理、病理过程中的作用,如发育、细胞分化、神经退化性疾病、压力条件下、感染和癌症。