HIC1 Rabbit pAb
HIC1 Rabbit pAb
- 产品详情
- 实验流程
- 背景知识
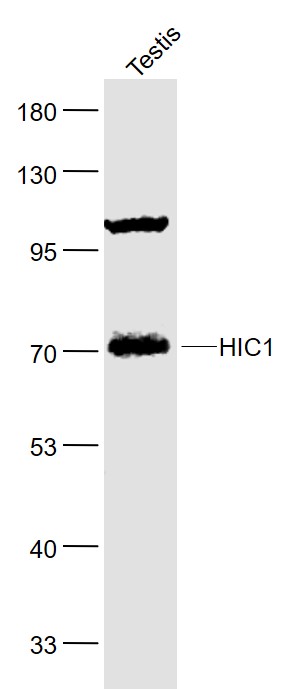
Application
| WB |
|---|---|
| Primary Accession | Q14526 |
| Reactivity | Mouse |
| Predicted | Human, Rat, Chicken, Dog, Pig, Horse |
| Host | Rabbit |
| Clonality | Polyclonal |
| Calculated MW | 76508 Da |
| Physical State | Liquid |
| Immunogen | KLH conjugated synthetic peptide derived from human HIC1 |
| Epitope Specificity | 501-650/733 |
| Isotype | IgG |
| Purity | affinity purified by Protein A |
| Buffer | 0.01M TBS (pH7.4) with 1% BSA, 0.02% Proclin300 and 50% Glycerol. |
| SUBCELLULAR LOCATION | Nucleus. |
| SIMILARITY | Belongs to the krueppel C2H2-type zinc-finger protein family. Hic subfamily.Contains 1 BTB (POZ) domain.Contains 5 C2H2-type zinc fingers. |
| SUBUNIT | Self-associates. Interacts with HIC2. Interacts with CTBP1 and CTBP2. Interacts with TCF7L2 and ARID1A. Interacts with MTA1 and MBD3; indicative for an association with the NuRD complex. |
| Post-translational modifications | Acetylated on several residues, including Lys-333. Lys-333 is deacetylated by SIRT1.Sumoylated on Lys-333 by a PIAS family member, which enhances interaction with MTA1, positively regulates transcriptional repression activity and is enhanced by HDAC4. |
| Important Note | This product as supplied is intended for research use only, not for use in human, therapeutic or diagnostic applications. |
| Background Descriptions | Hypermethylated in cancer (HIC-1) was originally identified as a target of p53-induced gene expression. HIC-1 is deleted in the genetic disorder Miller-Dieker syndrome (MDS), and the expression of HIC-1 is also frequently suppressed in leukemia and various cancers due to the hypermethylation of specific DNA regions and the resulting transcriptional silencing. These and other studies indicate that HIC-1 acts as a putative tumor suppressor protein that mediates transcriptional repression. HIC-1 is ubiquitously expressed in adult tissues and its structure is defined by five zinc fingers and an N-terminal broad complex POZ (or BTB) domain. In several BTB/POZ containing proteins, including BCL-6 and the promyelocytic leukemia zinc-finger (PLZF) oncoprotein, this domain interacts with the SMRT/N-CoR-mSin3A HDAC complex and is directly involved in repressing and silencing gene transcription. When this domain is deleted, as with the oncogenic PLZF-RAR chimera of promyelocytic leukemias, this transcriptional repression is attenuated. Conversely, HIC-1 does not interact with components of the HDAC complex, suggesting that HIC-1-induced transcriptional repression is unassociated with the POZ/BTB domain. |
| Gene ID | 3090 |
|---|---|
| Other Names | Hypermethylated in cancer 1 protein, Hic-1, Zinc finger and BTB domain-containing protein 29, HIC1, ZBTB29 |
| Target/Specificity | Ubiquitously expressed with highest levels found in lung, colon, prostate, thymus, testis and ovary. Expression is absent or decreased in many tumor cells. |
| Dilution | WB=1:500-2000 |
| Storage | Store at -20 °C for one year. Avoid repeated freeze/thaw cycles. When reconstituted in sterile pH 7.4 0.01M PBS or diluent of antibody the antibody is stable for at least two weeks at 2-4 °C. |
| Name | HIC1 |
|---|---|
| Synonyms | ZBTB29 |
| Function | Transcriptional repressor (PubMed:12052894, PubMed:15231840). Recognizes and binds to the consensus sequence '5- [CG]NG[CG]GGGCA[CA]CC-3' (PubMed:15231840). May act as a tumor suppressor (PubMed:20154726). Involved in development of head, face, limbs and ventral body wall (By similarity). Involved in down- regulation of SIRT1 and thereby is involved in regulation of p53/TP53- dependent apoptotic DNA-damage responses (PubMed:16269335). The specific target gene promoter association seems to be depend on corepressors, such as CTBP1 or CTBP2 and MTA1 (PubMed:12052894, PubMed:20547755). In cooperation with MTA1 (indicative for an association with the NuRD complex) represses transcription from CCND1/cyclin-D1 and CDKN1C/p57Kip2 specifically in quiescent cells (PubMed:20547755). Involved in regulation of the Wnt signaling pathway probably by association with TCF7L2 and preventing TCF7L2 and CTNNB1 association with promoters of TCF-responsive genes (PubMed:16724116). Seems to repress transcription from E2F1 and ATOH1 which involves ARID1A, indicative for the participation of a distinct SWI/SNF-type chromatin-remodeling complex (PubMed:18347096, PubMed:19486893). Probably represses transcription of ACKR3, FGFBP1 and EFNA1 (PubMed:16690027, PubMed:19525223, PubMed:20154726). |
| Cellular Location | Nucleus. |
| Tissue Location | Ubiquitously expressed with highest levels found in lung, colon, prostate, thymus, testis and ovary. Expression is absent or decreased in many tumor cells |
For Research Use Only. Not For Use In Diagnostic Procedures.
Provided below are standard protocols that you may find useful for product applications.
BACKGROUND
Hypermethylated in cancer (HIC-1) was originally identified as a target of p53-induced gene expression. HIC-1 is deleted in the genetic disorder Miller-Dieker syndrome (MDS), and the expression of HIC-1 is also frequently suppressed in leukemia and various cancers due to the hypermethylation of specific DNA regions and the resulting transcriptional silencing. These and other studies indicate that HIC-1 acts as a putative tumor suppressor protein that mediates transcriptional repression. HIC-1 is ubiquitously expressed in adult tissues and its structure is defined by five zinc fingers and an N-terminal broad complex POZ (or BTB) domain. In several BTB/POZ containing proteins, including BCL-6 and the promyelocytic leukemia zinc-finger (PLZF) oncoprotein, this domain interacts with the SMRT/N-CoR-mSin3A HDAC complex and is directly involved in repressing and silencing gene transcription. When this domain is deleted, as with the oncogenic PLZF-RAR chimera of promyelocytic leukemias, this transcriptional repression is attenuated. Conversely, HIC-1 does not interact with components of the HDAC complex, suggesting that HIC-1-induced transcriptional repression is unassociated with the POZ/BTB domain.
终于等到您。ABCEPTA(百远生物)抗体产品。
点击下方“我要评价 ”按钮提交您的反馈信息,您的反馈和评价是我们最宝贵的财富之一,
我们将在1-3个工作日内处理您的反馈信息。
如有疑问,联系:0512-88856768 tech-china@abcepta.com.






















 癌症的基本特征包括细胞增殖、血管生成、迁移、凋亡逃避机制和细胞永生等。找到癌症发生过程中这些通路的关键标记物和对应的抗体用于检测至关重要。
癌症的基本特征包括细胞增殖、血管生成、迁移、凋亡逃避机制和细胞永生等。找到癌症发生过程中这些通路的关键标记物和对应的抗体用于检测至关重要。 为您推荐一个泛素化位点预测神器——泛素化分析工具,可以为您的蛋白的泛素化位点作出预测和评分。
为您推荐一个泛素化位点预测神器——泛素化分析工具,可以为您的蛋白的泛素化位点作出预测和评分。 细胞自噬受体图形绘图工具为你的蛋白的细胞受体结合位点作出预测和评分,识别结合到自噬通路中的蛋白是非常重要的,便于让我们理解自噬在正常生理、病理过程中的作用,如发育、细胞分化、神经退化性疾病、压力条件下、感染和癌症。
细胞自噬受体图形绘图工具为你的蛋白的细胞受体结合位点作出预测和评分,识别结合到自噬通路中的蛋白是非常重要的,便于让我们理解自噬在正常生理、病理过程中的作用,如发育、细胞分化、神经退化性疾病、压力条件下、感染和癌症。