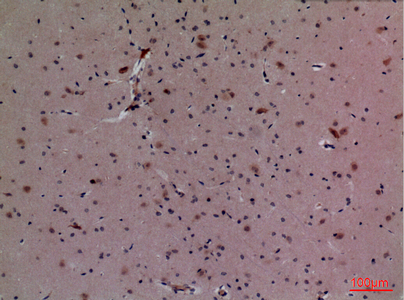
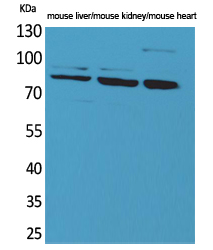
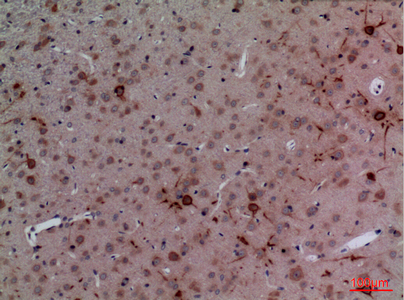
CUL-4B Polyclonal Antibody
- 产品详情
- 实验流程
- 背景知识
Application
| WB, IHC-P, IF, ICC, E |
|---|---|
| Primary Accession | Q13620 |
| Reactivity | Human, Mouse, Rat |
| Host | Rabbit |
| Clonality | Polyclonal |
| Calculated MW | 103982 Da |
| Gene ID | 8450 |
|---|---|
| Other Names | CUL4B; KIAA0695; Cullin-4B; CUL-4B |
| Dilution | WB~~Western Blot: 1/500 - 1/2000. IHC-p: 1:100-300 ELISA: 1/20000. Not yet tested in other applications. IHC-P~~Western Blot: 1/500 - 1/2000. IHC-p: 1:100-300 ELISA: 1/20000. Not yet tested in other applications. IF~~1:50~200 ICC~~N/A E~~N/A |
| Format | Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.09% (W/V) sodium azide. |
| Storage Conditions | -20℃ |
| Name | CUL4B {ECO:0000303|PubMed:14578910, ECO:0000312|HGNC:HGNC:2555} |
|---|---|
| Function | Core component of multiple cullin-RING-based E3 ubiquitin- protein ligase complexes which mediate the ubiquitination and subsequent proteasomal degradation of target proteins (PubMed:14578910, PubMed:16322693, PubMed:16678110, PubMed:18593899, PubMed:22118460, PubMed:29779948, PubMed:30166453, PubMed:33854232, PubMed:33854239). The functional specificity of the E3 ubiquitin-protein ligase complex depends on the variable substrate recognition subunit (PubMed:14578910, PubMed:16678110, PubMed:18593899, PubMed:22118460, PubMed:29779948). CUL4B may act within the complex as a scaffold protein, contributing to catalysis through positioning of the substrate and the ubiquitin- conjugating enzyme (PubMed:14578910, PubMed:16678110, PubMed:18593899, PubMed:22118460). Plays a role as part of the E3 ubiquitin-protein ligase complex in polyubiquitination of CDT1, histone H2A, histone H3 and histone H4 in response to radiation-induced DNA damage (PubMed:14578910, PubMed:16678110, PubMed:18593899). Targeted to UV damaged chromatin by DDB2 and may be important for DNA repair and DNA replication (PubMed:16678110). A number of DCX complexes (containing either TRPC4AP or DCAF12 as substrate-recognition component) are part of the DesCEND (destruction via C-end degrons) pathway, which recognizes a C-degron located at the extreme C terminus of target proteins, leading to their ubiquitination and degradation (PubMed:29779948). The DCX(AMBRA1) complex is a master regulator of the transition from G1 to S cell phase by mediating ubiquitination of phosphorylated cyclin-D (CCND1, CCND2 and CCND3) (PubMed:33854232, PubMed:33854239). The DCX(AMBRA1) complex also acts as a regulator of Cul5-RING (CRL5) E3 ubiquitin-protein ligase complexes by mediating ubiquitination and degradation of Elongin-C (ELOC) component of CRL5 complexes (PubMed:30166453). Required for ubiquitination of cyclin E (CCNE1 or CCNE2), and consequently, normal G1 cell cycle progression (PubMed:16322693, PubMed:19801544). Regulates the mammalian target-of- rapamycin (mTOR) pathway involved in control of cell growth, size and metabolism (PubMed:18235224). Specific CUL4B regulation of the mTORC1- mediated pathway is dependent upon 26S proteasome function and requires interaction between CUL4B and MLST8 (PubMed:18235224). With CUL4A, contributes to ribosome biogenesis (PubMed:26711351). |
| Cellular Location | Cytoplasm {ECO:0000250|UniProtKB:A2A432}. Nucleus. Note=More concentrated in nuclei than in cytoplasm in germinal vesicle (GV) stage oocytes, zygotes and the 2-cell stage, but distributed in the cytoplasm at the MII-stage oocytes. {ECO:0000250|UniProtKB:A2A432} |
For Research Use Only. Not For Use In Diagnostic Procedures.
Provided below are standard protocols that you may find useful for product applications.
BACKGROUND
Core component of multiple cullin-RING-based E3 ubiquitin-protein ligase complexes which mediate the ubiquitination and subsequent proteasomal degradation of target proteins. The functional specificity of the E3 ubiquitin-protein ligase complex depends on the variable substrate recognition subunit. CUL4B may act within the complex as a scaffold protein, contributing to catalysis through positioning of the substrate and the ubiquitin-conjugating enzyme. Plays a role as part of the E3 ubiquitin-protein ligase complex in polyubiquitination of CDT1, histone H2A, histone H3 and histone H4 in response to radiation- induced DNA damage. Targeted to UV damaged chromatin by DDB2 and may be important for DNA repair and DNA replication. Required for ubiquitination of cyclin E, and consequently, normal G1 cell cycle progression. Regulates the mammalian target-of-rapamycin (mTOR) pathway involved in control of cell growth, size and metabolism. Specific CUL4B regulation of the mTORC1-mediated pathway is dependent upon 26S proteasome function and requires interaction between CUL4B and MLST8.
终于等到您。ABCEPTA(百远生物)抗体产品。
点击下方“我要评价 ”按钮提交您的反馈信息,您的反馈和评价是我们最宝贵的财富之一,
我们将在1-3个工作日内处理您的反馈信息。
如有疑问,联系:0512-88856768 tech-china@abcepta.com.























 癌症的基本特征包括细胞增殖、血管生成、迁移、凋亡逃避机制和细胞永生等。找到癌症发生过程中这些通路的关键标记物和对应的抗体用于检测至关重要。
癌症的基本特征包括细胞增殖、血管生成、迁移、凋亡逃避机制和细胞永生等。找到癌症发生过程中这些通路的关键标记物和对应的抗体用于检测至关重要。 为您推荐一个泛素化位点预测神器——泛素化分析工具,可以为您的蛋白的泛素化位点作出预测和评分。
为您推荐一个泛素化位点预测神器——泛素化分析工具,可以为您的蛋白的泛素化位点作出预测和评分。 细胞自噬受体图形绘图工具为你的蛋白的细胞受体结合位点作出预测和评分,识别结合到自噬通路中的蛋白是非常重要的,便于让我们理解自噬在正常生理、病理过程中的作用,如发育、细胞分化、神经退化性疾病、压力条件下、感染和癌症。
细胞自噬受体图形绘图工具为你的蛋白的细胞受体结合位点作出预测和评分,识别结合到自噬通路中的蛋白是非常重要的,便于让我们理解自噬在正常生理、病理过程中的作用,如发育、细胞分化、神经退化性疾病、压力条件下、感染和癌症。
.jpg)
.jpg)
.jpg)