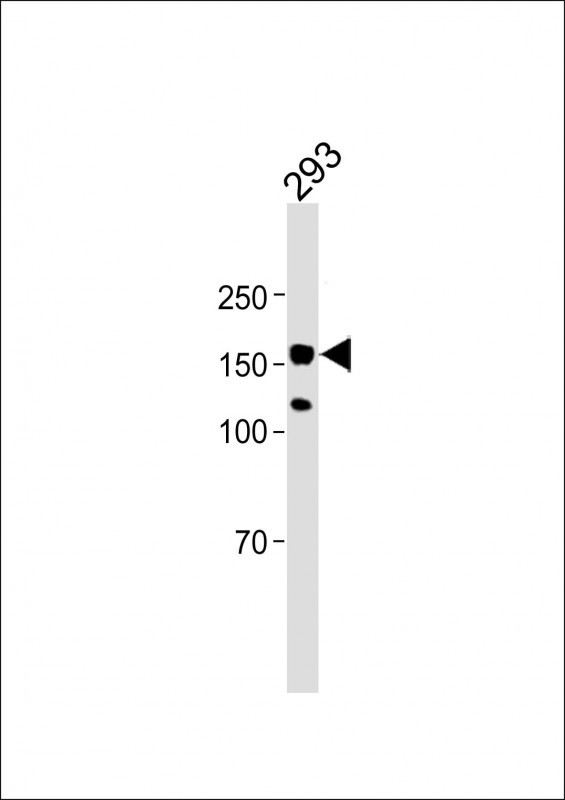
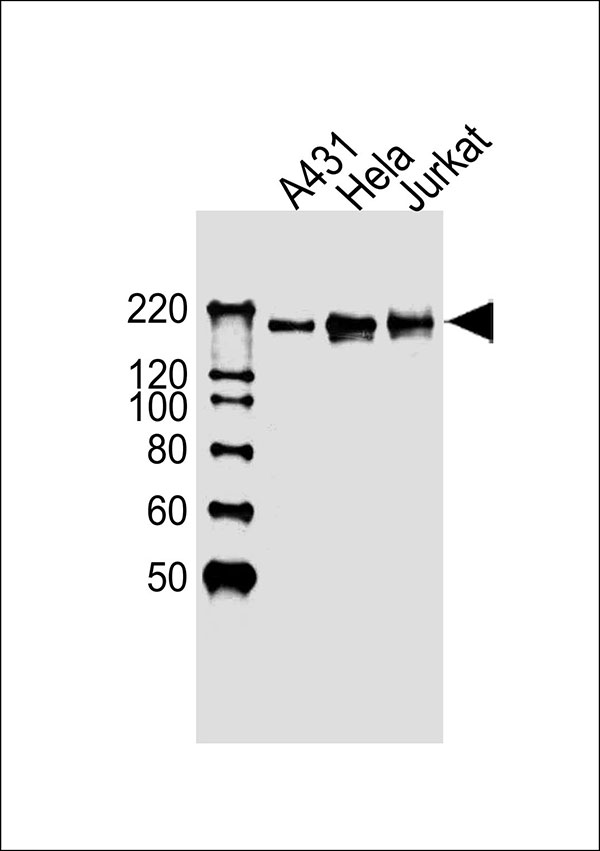
EHMT2 Antibody (Center)
Purified Rabbit Polyclonal Antibody (Pab)
- 产品详情
- 实验流程
- 背景知识
Application
| WB |
|---|---|
| Primary Accession | Q96KQ7 |
| Other Accession | Q9Z148 |
| Reactivity | Human |
| Predicted | Mouse, Rat |
| Host | Rabbit |
| Clonality | Polyclonal |
| Calculated MW | 132370 Da |
| Isotype | Rabbit IgG |
| Antigen Source | HUMAN |
| Gene ID | 10919 |
|---|---|
| Antigen Region | 361-395 aa |
| Other Names | Histone-lysine N-methyltransferase EHMT2, 211-, Euchromatic histone-lysine N-methyltransferase 2, HLA-B-associated transcript 8, Histone H3-K9 methyltransferase 3, H3-K9-HMTase 3, Lysine N-methyltransferase 1C, Protein G9a, EHMT2, BAT8, C6orf30, G9A, KMT1C, NG36 |
| Dilution | WB~~1:1000 |
| Target/Specificity | This EHMT2 antibody is generated from a rabbit immunized with a KLH conjugated synthetic peptide between 361-395 amino acids from the Central region of human EHMT2. |
| Format | Purified polyclonal antibody supplied in PBS with 0.05% (V/V) Proclin 300. This antibody is purified through a protein A column, followed by peptide affinity purification. |
| Storage | Maintain refrigerated at 2-8°C for up to 2 weeks. For long term storage store at -20°C in small aliquots to prevent freeze-thaw cycles. |
| Precautions | EHMT2 Antibody (Center) is for research use only and not for use in diagnostic or therapeutic procedures. |
| Name | EHMT2 |
|---|---|
| Synonyms | BAT8, C6orf30, G9A, KMT1C, NG36 |
| Function | Histone methyltransferase that specifically mono- and dimethylates 'Lys-9' of histone H3 (H3K9me1 and H3K9me2, respectively) in euchromatin. H3K9me represents a specific tag for epigenetic transcriptional repression by recruiting HP1 proteins to methylated histones (PubMed:11316813, PubMed:20084102). Also mediates monomethylation of 'Lys-56' of histone H3 (H3K56me1) in G1 phase, leading to promote interaction between histone H3 and PCNA and regulating DNA replication (PubMed:22387026). Also weakly methylates 'Lys-27' of histone H3 (H3K27me) (PubMed:11316813). Also required for DNA methylation, the histone methyltransferase activity is not required for DNA methylation, suggesting that these 2 activities function independently. Probably targeted to histone H3 by different DNA-binding proteins like E2F6, MGA, MAX and/or DP1. Also able to mono- and dimethylate histone H1-4 at 'Lys-26' (H1.4K26me1 and H1.4K26me2, respectively) (PubMed:19144645). In addition to the histone methyltransferase activity, also methylates non-histone proteins: mediates dimethylation of 'Lys-373' of p53/TP53 (PubMed:20118233). Also methylates CDYL, WIZ, ACIN1, DNMT1, HDAC1, ERCC6, KLF12 and itself (PubMed:18438403). |
| Cellular Location | Nucleus. Chromosome. Note=Associates with euchromatic regions (PubMed:11316813). Does not associate with heterochromatin (PubMed:11316813). |
| Tissue Location | Expressed in all tissues examined, with high levels in fetal liver, thymus, lymph node, spleen and peripheral blood leukocytes and lower level in bone marrow |
For Research Use Only. Not For Use In Diagnostic Procedures.
Provided below are standard protocols that you may find useful for product applications.
BACKGROUND
Histone methyltransferase that specifically mono- and dimethylates 'Lys-9' of histone H3 (H3K9me1 and H3K9me2, respectively) in euchromatin. H3K9me represents a specific tag for epigenetic transcriptional repression by recruiting HP1 proteins to methylated histones. Also mediates monomethylation of 'Lys-56' of histone H3 (H3K56me1) in G1 phase, leading to promote interaction between histone H3 and PCNA and regulating DNA replication. Also weakly methylates 'Lys-27' of histone H3 (H3K27me). Also required for DNA methylation, the histone methyltransferase activity is not required for DNA methylation, suggesting that these 2 activities function independently. Probably targeted to histone H3 by different DNA-binding proteins like E2F6, MGA, MAX and/or DP1. May also methylate histone H1. In addition to the histone methyltransferase activity, also methylates non-histone proteins: mediates dimethylation of 'Lys- 373' of p53/TP53. Also methylates CDYL, WIZ, ACIN1, DNMT1, HDAC1, ERCC6, KLF12 and itself.
REFERENCES
Brown S.E.,et al.Mamm. Genome 12:916-924(2001).
Ota T.,et al.Nat. Genet. 36:40-45(2004).
Xie T.,et al.Genome Res. 13:2621-2636(2003).
Mural R.J.,et al.Submitted (JUL-2005) to the EMBL/GenBank/DDBJ databases.
Hirakawa M.,et al.Submitted (SEP-1999) to the EMBL/GenBank/DDBJ databases.
终于等到您。ABCEPTA(百远生物)抗体产品。
点击下方“我要评价 ”按钮提交您的反馈信息,您的反馈和评价是我们最宝贵的财富之一,
我们将在1-3个工作日内处理您的反馈信息。
如有疑问,联系:0512-88856768 tech-china@abcepta.com.























 癌症的基本特征包括细胞增殖、血管生成、迁移、凋亡逃避机制和细胞永生等。找到癌症发生过程中这些通路的关键标记物和对应的抗体用于检测至关重要。
癌症的基本特征包括细胞增殖、血管生成、迁移、凋亡逃避机制和细胞永生等。找到癌症发生过程中这些通路的关键标记物和对应的抗体用于检测至关重要。 为您推荐一个泛素化位点预测神器——泛素化分析工具,可以为您的蛋白的泛素化位点作出预测和评分。
为您推荐一个泛素化位点预测神器——泛素化分析工具,可以为您的蛋白的泛素化位点作出预测和评分。 细胞自噬受体图形绘图工具为你的蛋白的细胞受体结合位点作出预测和评分,识别结合到自噬通路中的蛋白是非常重要的,便于让我们理解自噬在正常生理、病理过程中的作用,如发育、细胞分化、神经退化性疾病、压力条件下、感染和癌症。
细胞自噬受体图形绘图工具为你的蛋白的细胞受体结合位点作出预测和评分,识别结合到自噬通路中的蛋白是非常重要的,便于让我们理解自噬在正常生理、病理过程中的作用,如发育、细胞分化、神经退化性疾病、压力条件下、感染和癌症。